SELFIES Language Extension for VS Code
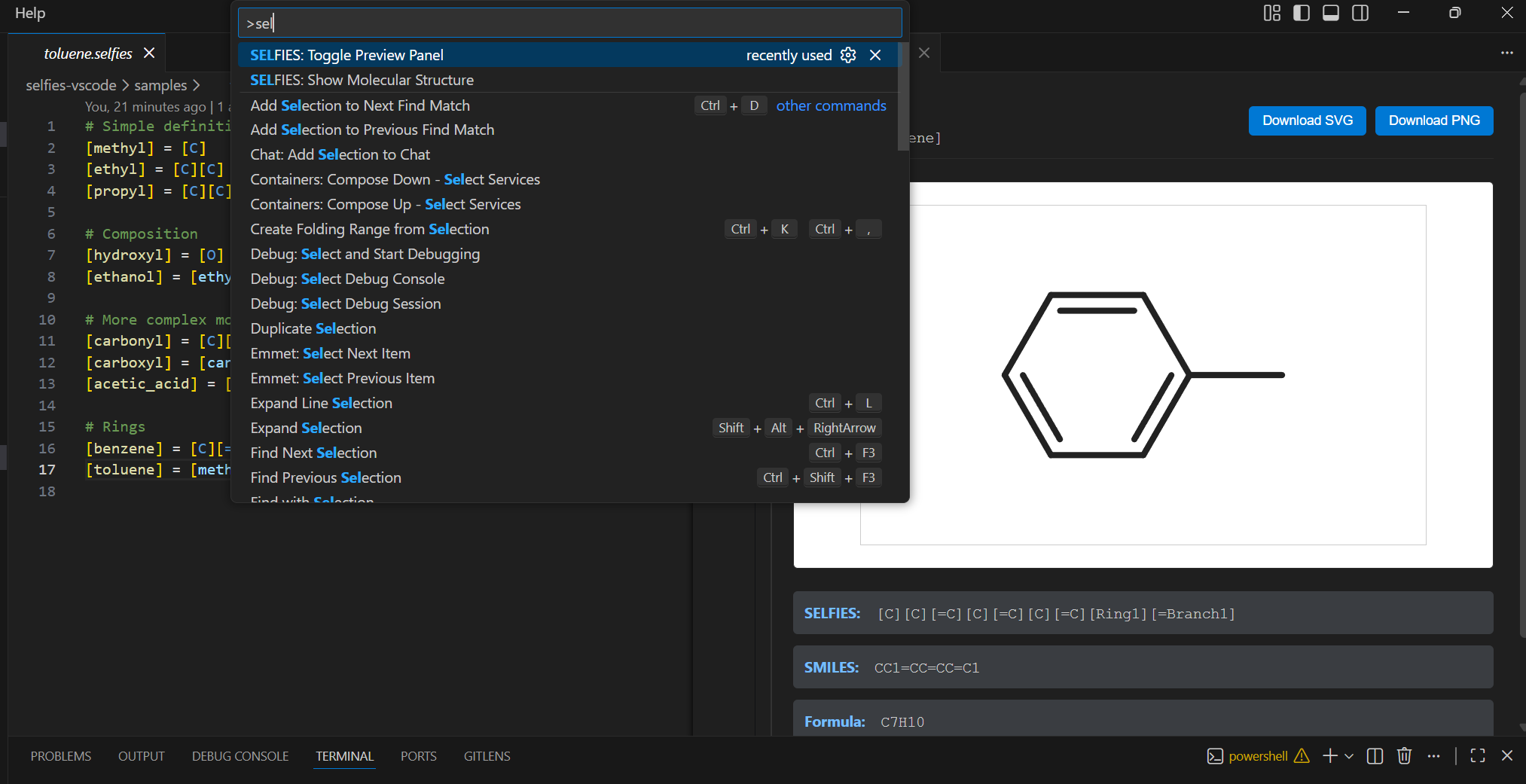
See your molecules as you write them.
Define molecular fragments, compose them into complex structures, and watch the visualization update live as you navigate your code.
Why SELFIES?
SELFIES was designed for machine learning — every string is a valid molecule, eliminating syntax errors in generative models. The DSL extends that principle: named fragments are easier for LLMs to compose correctly than raw atom strings, and undefined references fail loudly instead of producing silent errors.
The Problem
Pharmaceutical SMILES are write-only:
CC(=O)Nc1ccc(O)cc1
And when LLMs generate SMILES, they hallucinate invalid structures. SELFIES fixes the validity problem. The DSL fixes the readability problem.
With this extension, you write readable, composable definitions:
[acetyl] = [methyl][carbonyl]
[acetamide] = [acetyl][amino]
[acetaminophen] = [acetamide][para_hydroxyphenyl]
And see the structure instantly — formula, weight, 2D rendering — as your cursor moves through the file.
Installation
Install from VS Code Marketplace
Or search "SELFIES" in the VS Code extensions panel.
Quick Start
Using SELFIES DSL
- Create a file with
.selfies extension
- Start defining molecules — the preview panel opens automatically
- Move your cursor to any definition to see its structure
# fragments.selfies
[methyl] = [C]
[ethyl] = [C][C]
[hydroxyl] = [O]
[ethanol] = [ethyl][hydroxyl]
[methanol] = [methyl][hydroxyl]
Using smiles-js (JavaScript)
- Create a file with
.smiles.js extension
- Import Fragment and Ring from
smiles-js
- Export your molecules as constants
- Move your cursor to any exported molecule to see its structure
// molecules.smiles.js
import { Fragment, Ring } from 'smiles-js';
import { benzene, methyl, ethyl, hydroxyl } from 'smiles-js/common';
export const methane = Fragment('C');
export const ethanol = ethyl(hydroxyl);
export const toluene = benzene(methyl);
The extension executes your JavaScript file and reads the .smiles property from exported fragments - no regex parsing!
Features
SELFIES DSL (.selfies files)
- Domain-specific language for molecular composition
- Multi-file imports and reusable fragments
- Real-time diagnostics and error checking
smiles-js (.smiles.js files)
- JavaScript-based molecular construction
- Use Fragment() and Ring() functions
- Full JavaScript IDE support with molecular previews
Live Preview
As your cursor moves, the preview panel shows:
- 2D molecular structure
- Molecular formula
- Molecular weight
- SMILES output
- Export as SVG or PNG
Works for both .selfies and .smiles.js files!
Syntax Highlighting
Full highlighting for atoms, bonds, branches, rings, comments, and references in SELFIES files. JavaScript syntax highlighting for .smiles.js files.
Real-time Diagnostics
Instant feedback on SELFIES files:
- Undefined references
- Circular dependencies
- Duplicate definitions
- Syntax errors
Multi-file Projects
Import fragments across SELFIES files:
import "./fragments.selfies" # import all
import [methyl, ethyl] from "./fragments.selfies" # import specific
Or use standard JavaScript imports in .smiles.js files:
import { Fragment } from 'smiles-js';
import { benzene, methyl } from 'smiles-js/common';
Commands
SELFIES: Show Molecular Structure — Open the preview panelSELFIES: Toggle Preview Panel — Toggle preview on/offSELFIES: Refactor Molecule to Code — Convert a Fragment to constructor code (right-click menu in .smiles.js files)
Refactor to Code (Experimental)
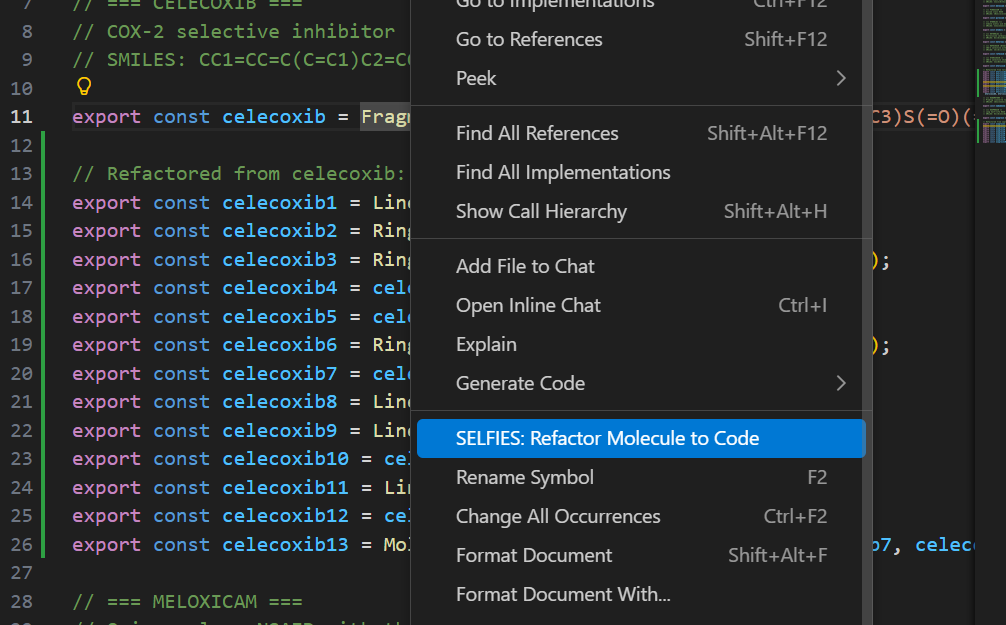
Right-click on any line with an exported Fragment in a .smiles.js file and select "SELFIES: Refactor Molecule to Code" to convert it into explicit Ring/Linear constructor calls.
Note: This feature is early in development and often fails for complex molecular structures. It works best with simple fragments.
Settings
| Setting |
Default |
Description |
selfies.previewOnCursorMove |
true |
Update preview when cursor moves |
selfies.autoOpenPreview |
true |
Auto-open preview for .selfies and .smiles.js files |
selfies.renderingEngine |
smiles-drawer |
Molecule rendering engine |
Language Rules
- No forward references — Define before you use
- No recursion — No circular dependencies
- Single assignment — No redefinitions
- Case sensitive —
[Methyl] ≠ [methyl]
- selfies-js — SELFIES DSL library (also usable as CLI and npm package)
- smiles-js — JavaScript library for composable molecular fragments
- SELFIES paper — Original research by the Aspuru-Guzik group
Citation
If you use SELFIES in your research:
Krenn, M., Häse, F., Nigam, A., Friederich, P., & Aspuru-Guzik, A. (2020). Self-referencing embedded strings (SELFIES): A 100% robust molecular string representation. Machine Learning: Science and Technology, 1(4), 045024.
License
MIT



