
BioNetGen VS Code extension
This is a VS Code language extension for BioNetGen modeling language. Please read the installation instructions and see here for a starter guide.
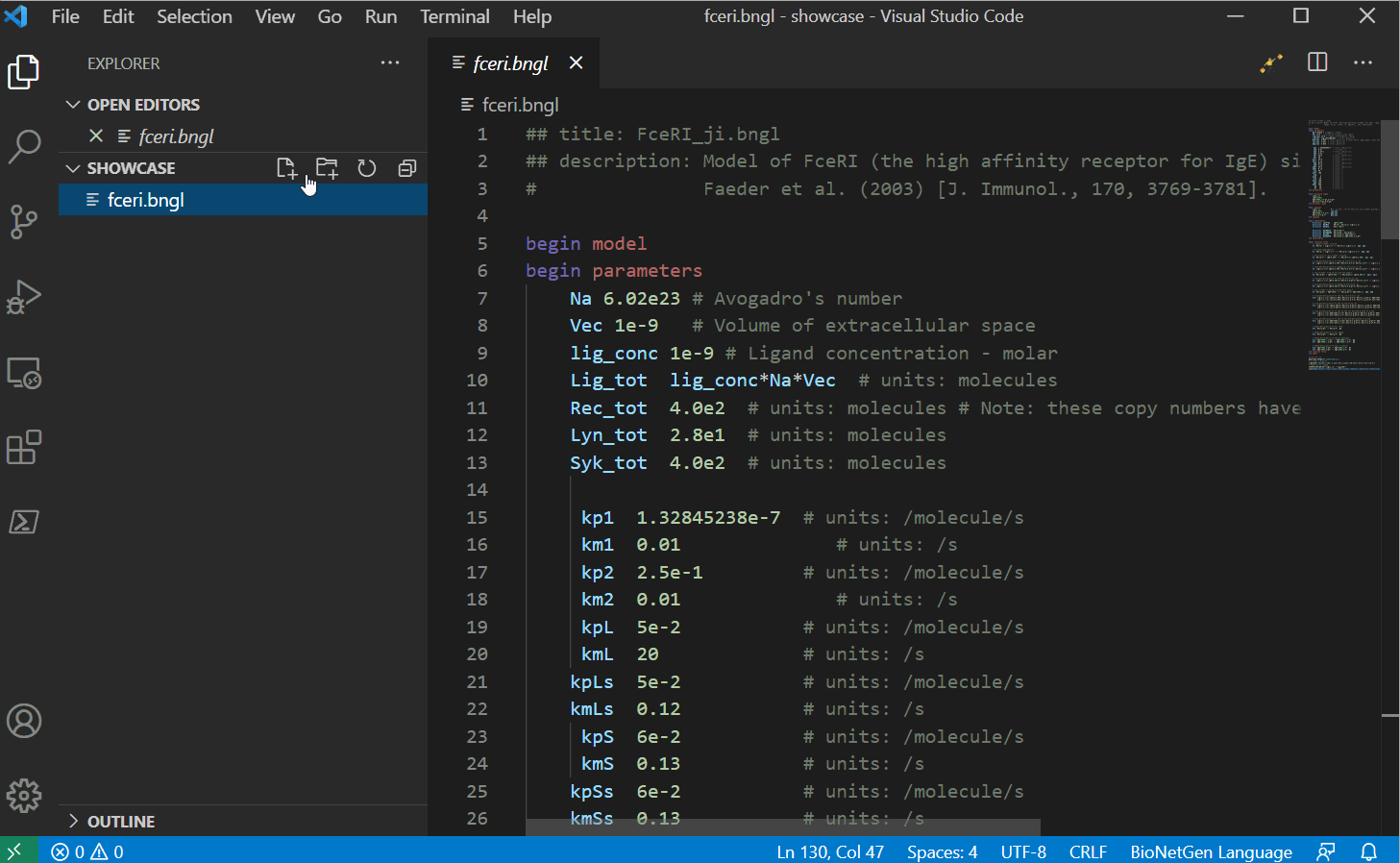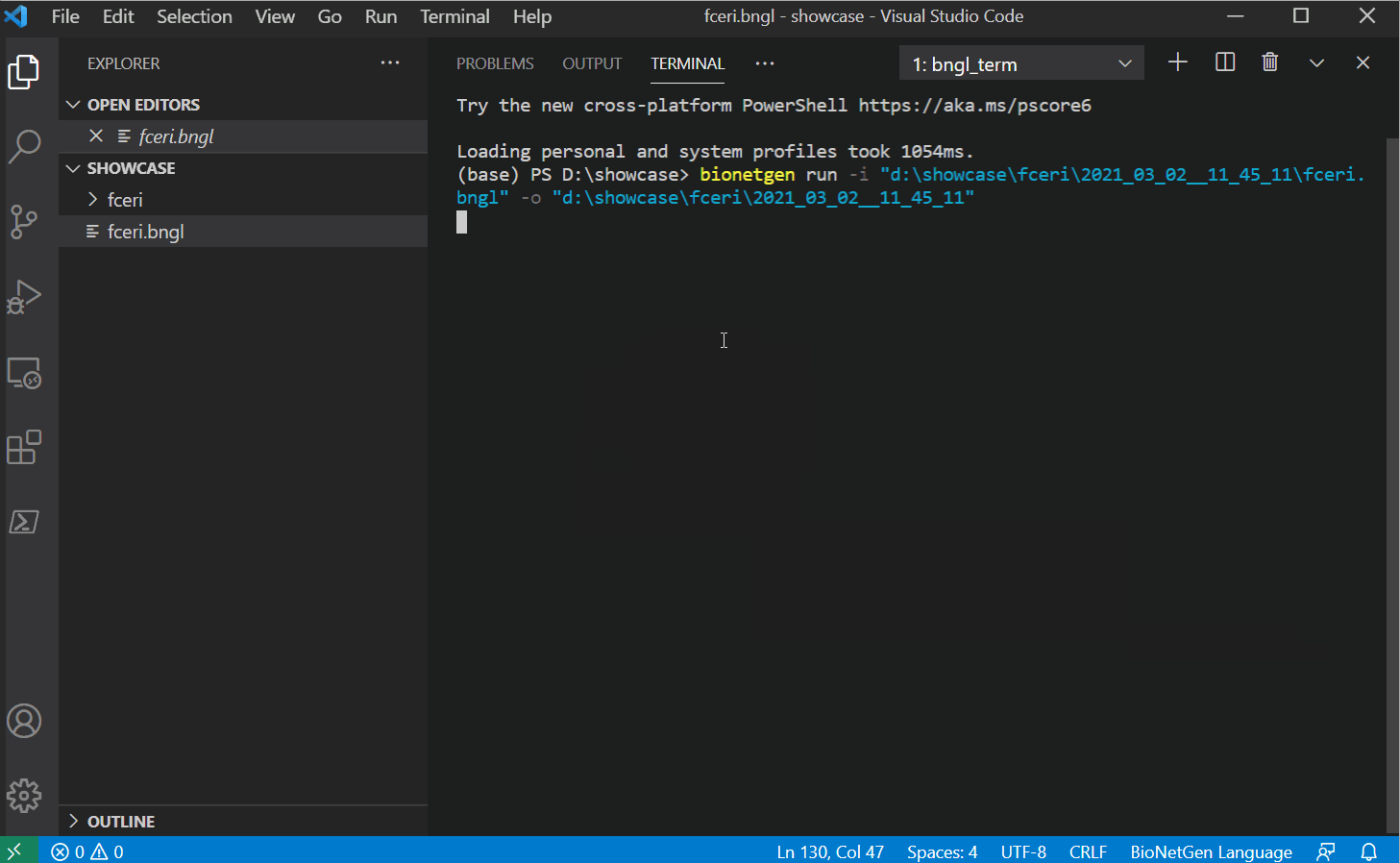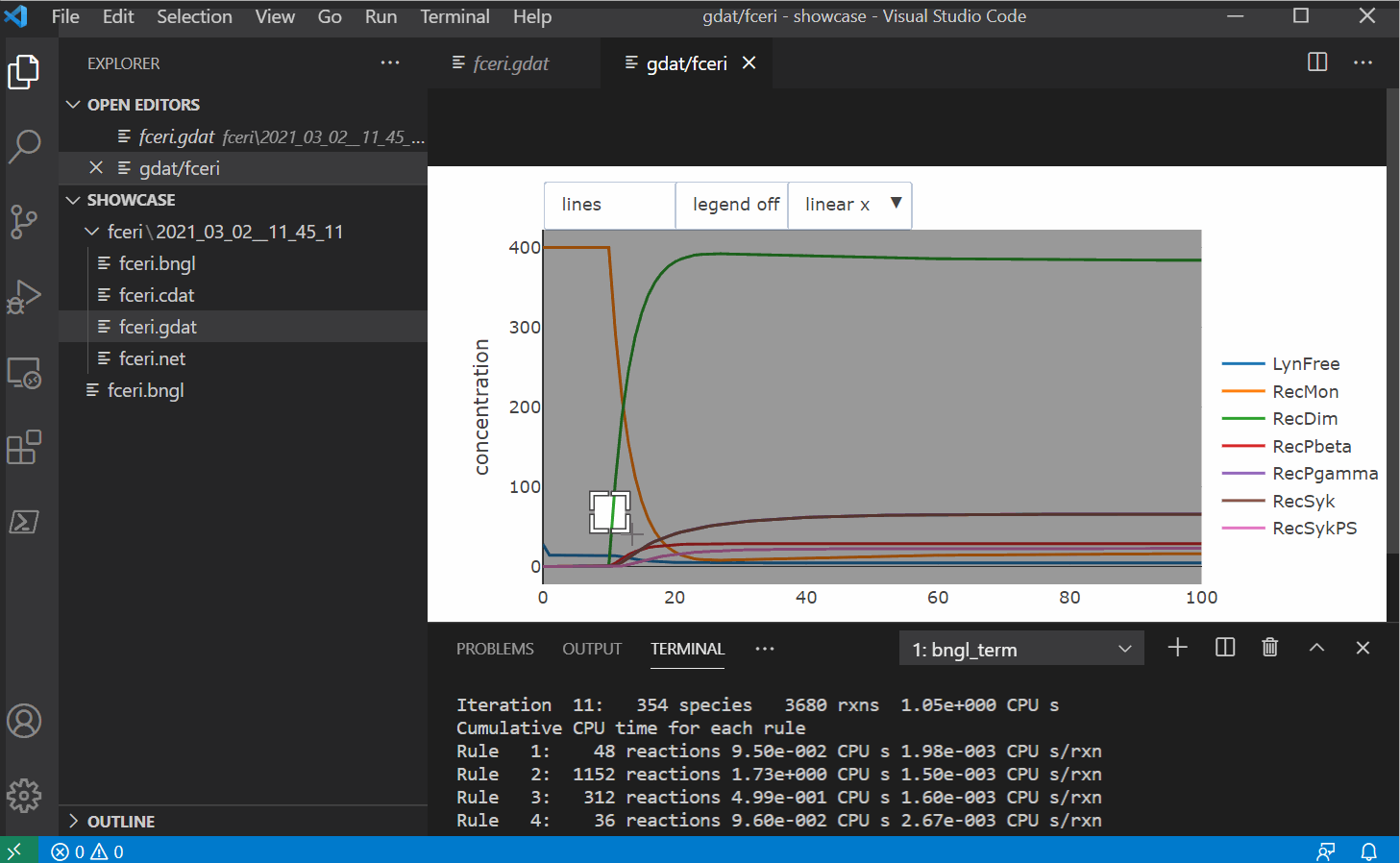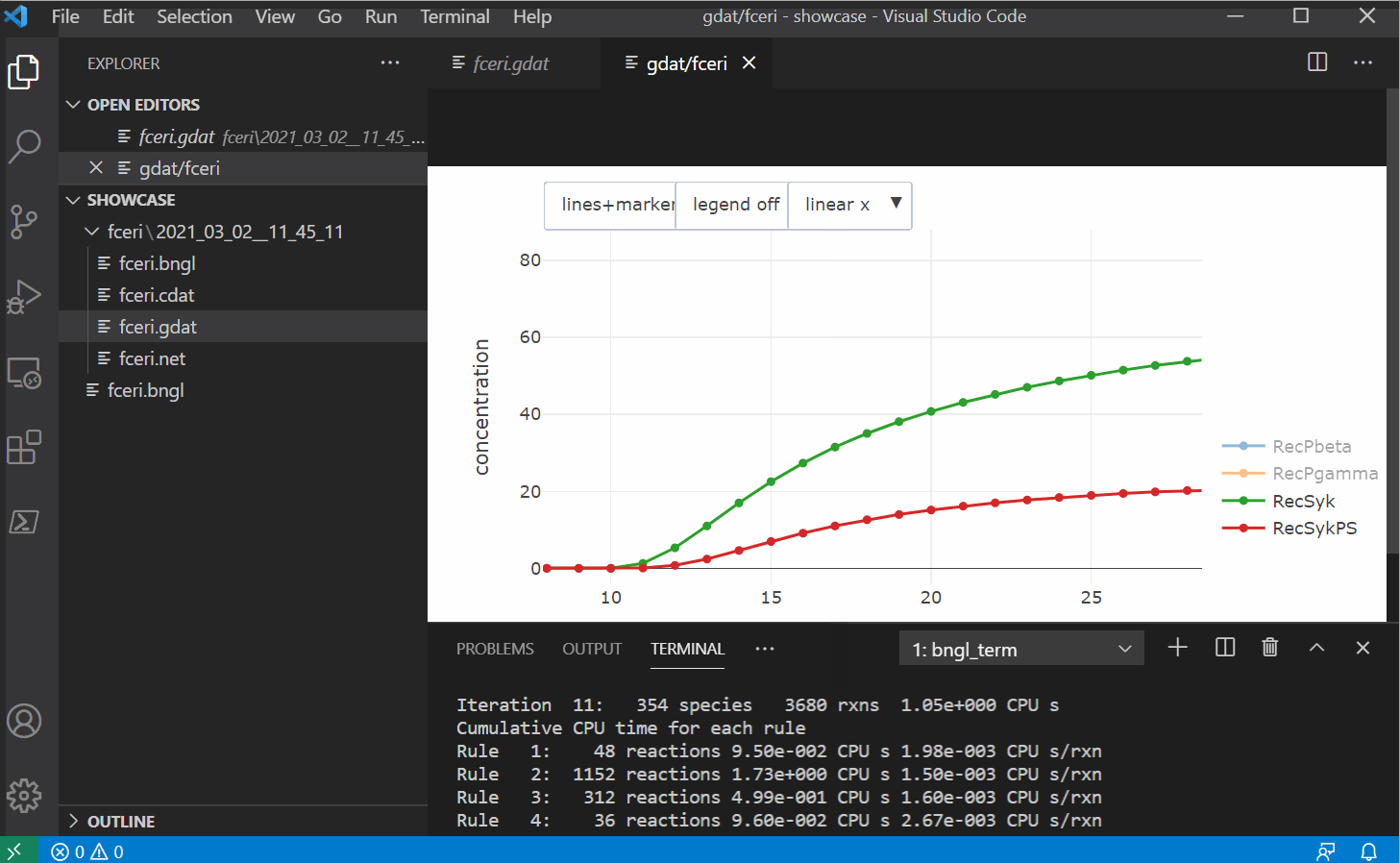
Features
- Syntax highlighting for BioNetGen modelling language
- Various snippets to make writing BNGL simpler
- A run button that automatically generates a timestamped folder and runs the current model
- A plot button that generates a plot of the current .gdat/.cdat/.scan file using the CLI
- Another plotting button that uses plotly to generate built-in plots within VS Code
Requirements
To use the run and plot buttons the default VS Code terminal you are using needs to have access to
Please note that this tool is in active early development and is subject to sweeping changes.
Installation
The extension can be found in the VS Code marketplace as BioNetGen Language. See here to learn how to browse and install extensions in VS Code.
For other ways to install, check out the installation guide.
Tips:
- Make sure to select a folder when opening up your model, currently that's how BNG extension knows where to save your results.
- For correct highlighting, please make sure
dark-bngl theme is selected, you can see how to change themes here
- If suggestions disappear for snippets, you can press
CTRL/CMD + space to show suggestions again
Known Issues
General issues
- Need a
default results folder to save results in case a folder wasn't selected before opening up a bngl file.
Highlighting issues
- Using line breaks disrupt syntax highlighting
Plotting issues
- Built-in plotting button alignment is not automated
- Sometimes the plot doesn't show up the first time you click the button
Please submit an issue here if you find one or have any feature requests.
Release Notes
This extension is still in alpha stage of development.
| |


