NiiVue for VS Code
WebGL 2.0 medical image viewer for Visual Studio Code
A VS Code extension that uses NiiVue to display neuroimaging files directly in VS Code. View NIfTI files, meshes, tractography, and DICOM images with an interactive, hardware-accelerated viewer. Works seamlessly in remote sessions (SSH, WSL, containers) - especially useful for analyzing data on clusters and servers.
Installation
Install from the VS Code Marketplace:
- Open VS Code Extensions (
Ctrl+Shift+X)
- Search for "niivue" and install
Or install from the command line:
code --install-extension KorbinianEckstein.niivue
Usage
Opening Files
- Explorer: Simply click any supported neuroimaging file in the VS Code Explorer
- Compare Multiple Files: Select multiple files in Explorer, right-click → "NiiVue: Compare"
- Add Overlays: Click the Overlay menu
Remote Development
NiiVue works with VS Code's remote development features:
- SSH: View images on remote servers without downloading
- WSL: Access Windows Subsystem for Linux files
- Containers: Work with images inside Docker containers
- Codespaces: Use in GitHub Codespaces environments
- Web Version: Works in vscode.dev and github.dev
Screenshots
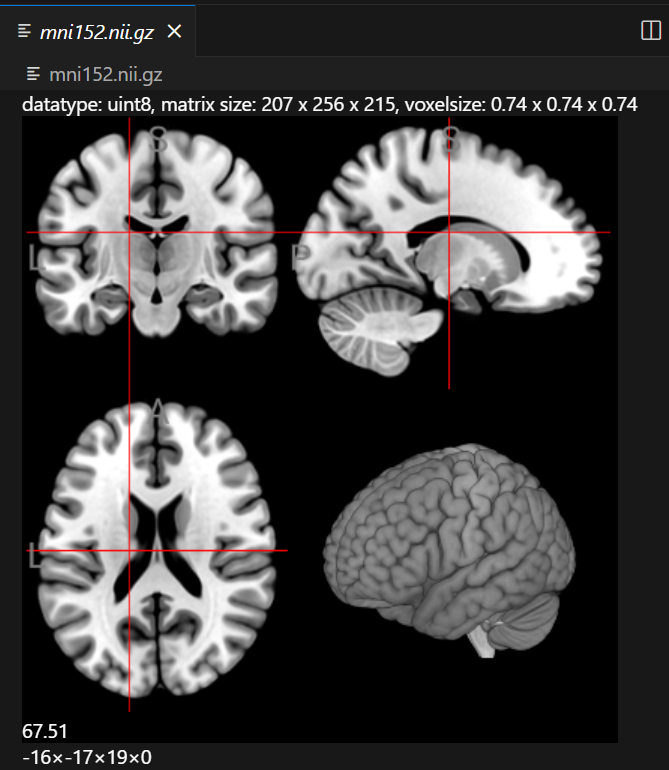
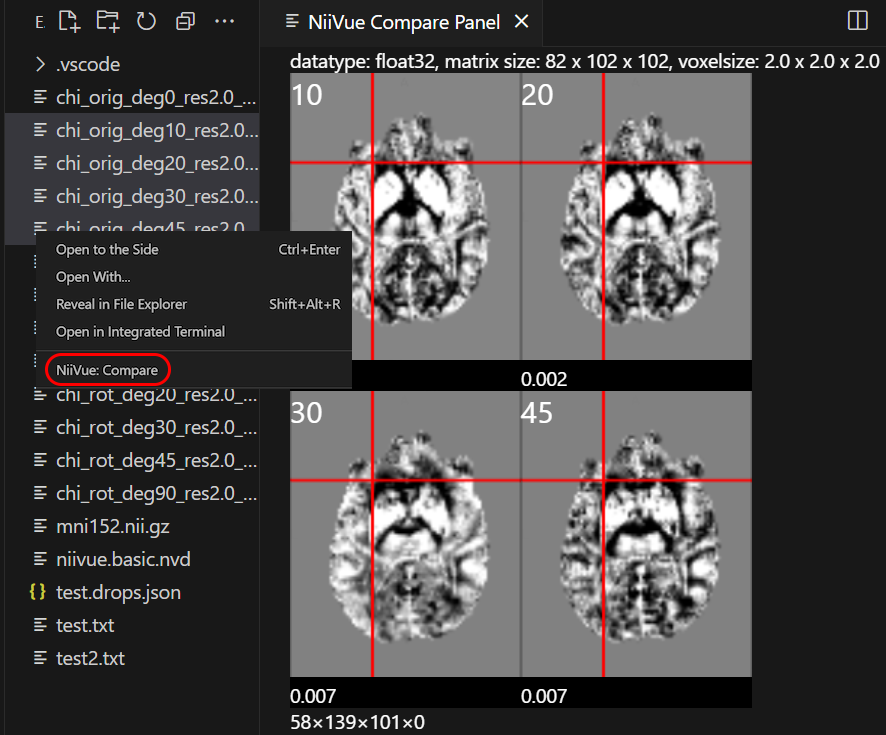
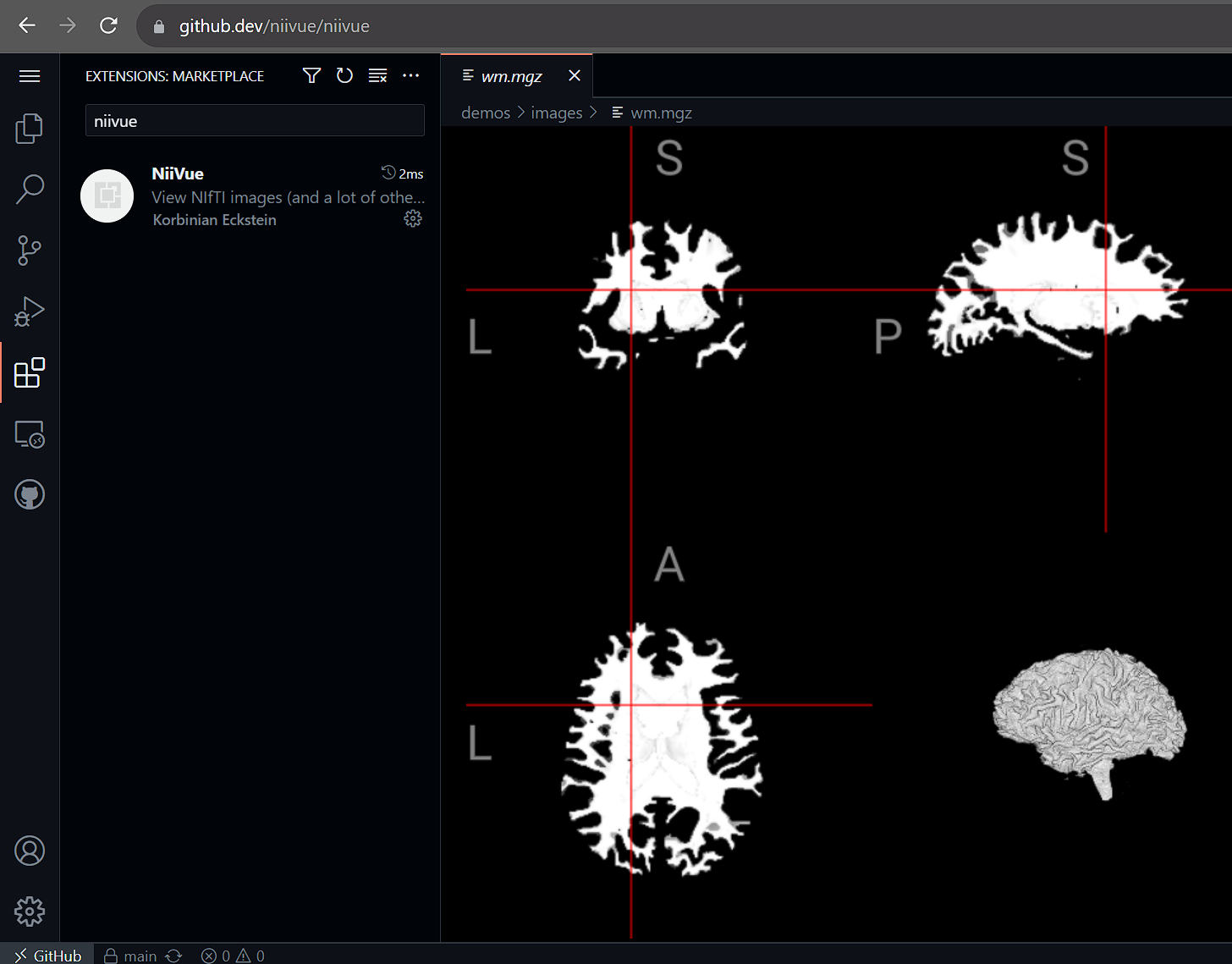
Keyboard Shortcuts
All keyboard shortcuts are displayed in menus and button tooltips throughout the interface. Hover over any button or menu item to see its shortcut.
Mouse Controls
- Right Mouse: Adjust contrast/brightness (windowing); with Zoom button pressed: drag to zoom
- Middle Mouse: Drag to pan
- Mouse Scroll: Change slice in currently hovered image
- Shift + Mouse: 2D dragging and 3D viewplane rotation
Core NiiVue Shortcuts (Built-in)
These shortcuts are handled by the niivue.js library and cannot be customized:
Navigation:
- ← →: Change volume in 4D images
- V: Cycle through view modes
- C: Cycle through clip plane orientations in 3D render
Crosshair Movement:
- H: Move crosshair to R (Right)
- L: Move crosshair to L (Left)
- J: Move crosshair to P (Posterior)
- K: Move crosshair to A (Anterior)
- Ctrl+U: Move crosshair to S (Superior)
- Ctrl+D: Move crosshair to I (Inferior)
UI Shortcuts (Customizable)
These shortcuts can be customized in VS Code's Keyboard Shortcuts editor (File → Preferences → Keyboard Shortcuts, search for "NiiVue"):
View Modes:
- 1: Axial view
- 2: Sagittal view
- 3: Coronal view
- 4: Render view
- 5: Multiplanar + Render view
- R: Reset view/zoom
Display Toggles:
- I: Toggle interpolation
- B: Toggle colorbar
- X: Toggle radiological convention
- M: Toggle crosshair visibility
- Z: Toggle zoom drag mode
- U: Cycle UI visibility (Show All → Hide UI → Hide All)
Actions:
- Ctrl+O (Cmd+O on Mac): Add image
- Ctrl+L (Cmd+L on Mac): Add overlay
- S: Open colorscale menu
- Ctrl+H (Cmd+H on Mac): Show header information
Tip: You can view all available shortcuts in the NiiVue menu bar at the top of the viewer, or by opening the VS Code Command Palette (Ctrl+Shift+P / Cmd+Shift+P) and typing "NiiVue".
- Voxel-based: NIfTI (.nii, .nii.gz), NRRD (.nrrd, .nhdr), MRtrix MIF (.mif), AFNI HEAD/BRIK, MGH/MGZ, ITK MHD (.mhd, .mha), ECAT7 (.v), DICOM (.dcm)
- Mesh-based: GIfTI (.gii), FreeSurfer (pial, white, inflated), MZ3 (.mz3), STL (.stl), Wavefront OBJ (.obj), PLY (.ply), BrainSuite DFS (.dfs), Legacy VTK (.vtk), X3D (.x3d), and others (ASC, BYU, GEO, ICO, TRI, OFF, SRF, NV)
- Mesh Overlays: GIfTI (.gii), CIfTI-2 (.nii), MZ3 (.mz3), FreeSurfer (CURV, ANNOT), SMP, STC
- Tractography: TCK (.tck), TRK (.trk), TRX (.trx), VTK (.vtk), AFNI (.niml.tract)
Web Version
A standalone web version is also available that can be installed as a Progressive Web App with file associations (Chrome/Edge only).
Development
This extension is part of the niivue-vscode monorepo. Contributions are welcome!
For development instructions, see DEVELOPMENT.md.
If you encounter issues or have feature requests, please open an issue on GitHub.
Support This Project
If you find this extension useful, please consider supporting its development:

License
BSD-2-Clause
Credits
- Built with NiiVue
- Initial development at the Neurodesk team. Neurodesk is a flexible and scalable data analysis environment for reproducible neuroimaging





