CCView - Comp Chem Viewer
A VS Code extension for viewing and analyzing computational chemistry output files. CCView reads computational chemistry calculation results, parses them using the cclib library, and displays molecular structures in 3D using the miew molecular viewer - all within VS Code.
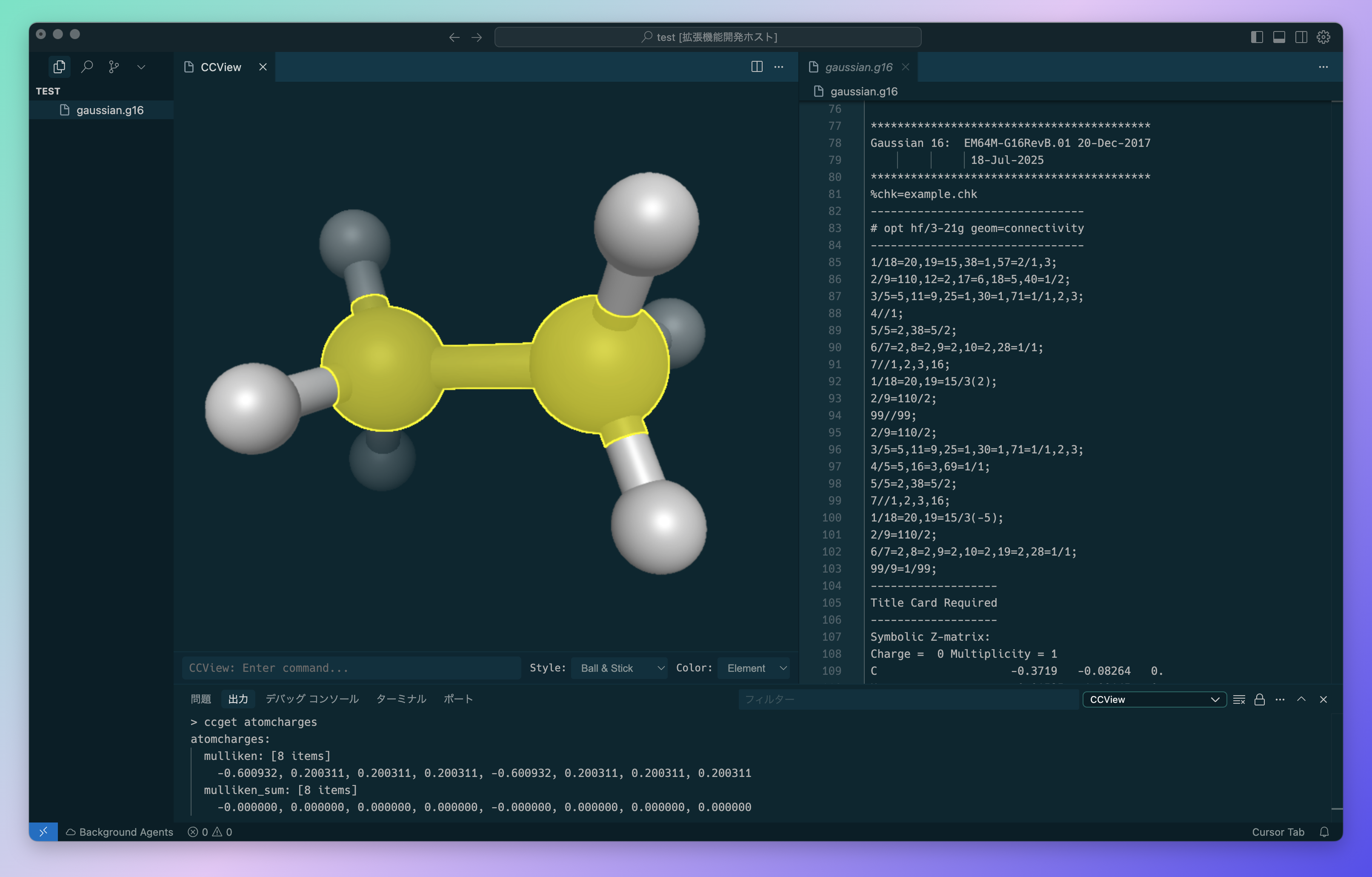
What CCView Does
CCView helps you work with computational chemistry files by:
- Reading calculation output files from computational chemistry software (Gaussian, ORCA, NWChem, etc.)
- Extracting molecular data using cclib - a library specifically designed for parsing computational chemistry files
- Displaying 3D molecular structures using miew - a web-based molecular viewer
- Providing analysis tools through an interactive command interface
The main advantage is that you can view your calculation results immediately after they complete, without leaving VS Code. Instead of opening separate programs, you can right-click on your output file and see the molecular structure directly in the editor.
Key Features
3D Molecular Visualization
- View molecular structures in 3D with rotation, zoom, and pan controls
- Support for different molecular representations (ball & stick, licorice, etc.)
- Works with both direct structure files (PDB, CIF, XYZ) and quantum chemistry output files
- Extract specific data from calculation results (energies, frequencies, atomic charges, etc.)
- Export molecular structures in various formats (XYZ, CML, JSON)
- View calculation properties through simple commands
Interactive Command Interface
- Built-in command interface for running analysis commands
- Command history and suggestions
- Direct access to cclib's ccget and ccwrite functionality
How It Works
For Computational Chemistry Output Files
- CCView uses cclib to read and parse your calculation output file
- Extracts molecular coordinates, energies, frequencies, and other data
- Displays the molecular structure in 3D using miew
- Provides access to the parsed data through analysis commands
For Direct Structure Files
- Loads PDB, CIF, or XYZ files directly into the 3D viewer
- No parsing required - immediate visualization
- Full miew functionality available
Computational Chemistry Software Output
CCView can read output files from most major computational chemistry programs:
| Software |
File Extensions |
Description |
| Gaussian |
.g09, .g16 |
Most widely used comp chem software |
| ORCA |
.orca, .nwo |
Free comp chem package |
| NWChem |
.nw |
Open source comp chem software |
| Psi4 |
.psi4 |
Open source comp chem software |
| GAMESS |
.gamess, .gms |
American and UK versions |
| Q-Chem |
.qchem |
Commercial comp chem software |
| Turbomole |
.tmol |
Commercial comp chem software |
| Molpro |
.molpro |
Commercial comp chem software |
| Molcas |
.molcas |
Commercial comp chem software |
| ADF |
.adf |
Amsterdam Density Functional |
| MOPAC |
.mopac |
Semi-empirical comp chem software |
| xTB |
.xtb |
Semi-empirical comp chem software |
| DALTON |
.dalton |
Open source comp chem software |
Direct Structure Files
| Format |
Extension |
Description |
| PDB |
.pdb |
Protein Data Bank format |
| CIF |
.cif |
Crystallographic Information Framework |
| XYZ |
.xyz |
Simple coordinate format |
Custom Extensions
You can also configure CCView to work with files using .log, .out, .txt, or .dat extensions. CCView will attempt to detect if these files contain computational chemistry output data.
Installation
Prerequisites
- VS Code (version 1.74.0 or later)
- Python (3.7 or later)
- VS Code Python Extension (recommended)
Install CCView
- Install CCView from the VS Code Marketplace
- Install required Python packages:
pip install cclib==1.8.1
Note: CCView is currently tested with cclib 1.8.1. While cclib 2.0 is in development, it may have compatibility issues. Please use the stable 1.8.1 version for now.
- Ensure VS Code can find your Python interpreter
Quick Start
- Open a supported file in VS Code (any computational chemistry output file or structure file)
- Right-click in the file explorer and select "Open CCView"
- View the 3D structure in the molecular viewer
- Use the command interface to analyze data (type
help to see available commands)
Using the Command Interface
CCView includes a command interface where you can run analysis commands. The command interface supports:
ccget Commands - Extract Data
ccget natom # Get number of atoms
ccget charge # Get total charge
ccget mult # Get multiplicity
ccget atomcharges # Get atomic charges
ccget scfenergies # Get SCF energies
ccget vibfreqs # Get vibrational frequencies
ccget vibirs # Get IR intensities
ccwrite Commands - Export Data
ccwrite xyz # Display in XYZ format
ccwrite xyz molecule.xyz # Save as XYZ file
ccwrite cml molecule.cml # Save as CML file
ccwrite json molecule.json # Save as JSON file
miew Commands - Control Visualization
miew rep 0 m=LC # Set representation to licorice
miew screenshot # Take a screenshot
miew set autoRotation 0.5 # Set auto rotation speed
miew help # Show miew help
Configuration
You can customize which file types CCView recognizes by editing VS Code settings:
- Open VS Code Settings (Ctrl/Cmd + ,)
- Search for "CCView"
- Modify the following settings:
File Type Settings
- Direct Formats: File extensions that load directly without parsing (default:
.pdb, .cif, .xyz)
- Chemistry Extensions: Known computational chemistry software extensions (default:
.g09, .g16, .orca, etc.)
- Custom Extensions: Additional extensions to process with cclib (default:
.log, .out, .txt, .dat)
Example Configuration
{
"ccview.directFormats": [".pdb", ".cif", ".xyz", ".mol"],
"ccview.chemistryExtensions": [".g09", ".g16", ".orca", ".nwo", ".psi4"],
"ccview.customExtensions": [".log", ".out", ".txt", ".dat", ".res"]
}
Common Use Cases
For Computational Chemists
- Quick structure verification after calculations complete
- Data extraction using ccget commands
- Format conversion for further analysis
- Structure comparison between different calculations
For Structural Biologists
- Protein structure viewing with different representations
- Secondary structure analysis with color coding
- Coordinate export in standard formats
- Interactive structure exploration
For Chemistry Educators
- Molecular visualization for teaching concepts
- Different representation modes (ball & stick, licorice, etc.)
- Interactive learning through analysis commands
- Structure export for presentations
Troubleshooting
Common Issues and Solutions
| Problem |
Solution |
| Python not found |
Install VS Code Python extension and select interpreter |
| cclib not installed |
Run pip install cclib==1.8.1 in your system terminal |
| File not recognized |
Check file extension and ensure file contains valid data |
| 3D viewer not loading |
Verify file contains valid molecular data |
Getting Help
- Check the command interface output for error messages
- Ensure your Python environment has the required packages
- Verify file format is supported
- Check VS Code's Developer Console for additional error information
Development
Building from Source
Clone the repository:
git clone https://github.com/yamnor/ccview.git
cd ccview
Install dependencies:
npm install
Compile and bundle:
npm run compile
npm run bundle
Press F5 in VS Code to launch in debug mode
Project Structure
src/ - TypeScript source code for VS Code extensionpython/ - Python backend for file parsingmedia/ - Bundled libraries (miew, etc.)webpack.config.js - Build configuration
Technical Details
Architecture
- Frontend: TypeScript VS Code extension with WebView
- Backend: Python with cclib for file parsing
- 3D Visualization: miew.js molecular viewer
Dependencies
- cclib: Computational chemistry file parsing
- miew: 3D molecular visualization
- Three.js: 3D graphics (via miew)
License
This project is licensed under the MIT License - see the LICENSE file for details.
Acknowledgments
- cclib - Computational chemistry file parsing library
- miew - 3D molecular visualization library
Support
CCView - Making computational chemistry accessible and interactive in VS Code



