Scikit-learn Snippets
This extension is for data scientists writing Python code to create, fit, and evaluate machine learning models using the scikit-learn package. It provides code snippets that work with Python (.py) files or (.ipynb) notebooks to ease the friction of working with scikit-learn. Snippets boost productivity by reducing the amount of Python syntax and function arguments that you need to remember, eliminating keystrokes, and helping to free the data scientist to focus on building useful models from their data.
1. Usage
1.1 Overview
All snippets provided by this extension have triggers prefixed sk, which activate the IntelliSense pop up or allow filtering in the Command Palette. Trigger prefixes are further organised as follows:
| Prefix |
Description |
sk-setup |
Starting point for importing commonly used modules and setting defaults. |
sk-read |
Read input training data or existing models from file. |
sk-prep |
Preprocess input training data for model fitting. |
sk-regress |
Create and fit regression models. |
sk-classify |
Create and fit classification models. |
sk-cluster |
Create and fit clustering models. |
sk-density |
Create and fit density estimation models. |
sk-embed |
Create and fit dimensionality reduction (embedding) models. |
sk-anomaly |
Create and fit anomaly detection models. |
sk-validation |
Model validation. |
sk-inspect |
Model inspection and explainability. |
sk-io |
Save and restore models on disk. |
sk-args |
Select and adjust model parameters from lists of valid options. |
For more information about the organization of the snippets provided by this extension see Section 4 for a visual reference of the complete hierarchy.
1.2 Snippets for machine learning workflows
A typical workflow is to:
Import commonly used modules with sk-setup,
Read training data from file with sk-read,
Preprocess training data with sk-prep,
Create and train models across a range of machine learning tasks such as:
regression (sk-regress),
classification (sk-classify),
clustering (sk-cluster),
density estimation (sk-density),
dimensionality reduction (sk-embed),
anomaly detection (sk-anomaly)
Evaluate fitted models by cross-validation (sk-validation) and inspection (sk-inspect),
Save and restore models on disk (sk-io),
Optionally adjust the parameters that control model fitting with sk-args.
See the Features section below for a full list of the available snippets and their prefix triggers.
1.3 Inserting snippets
Inserting machine learning code snippets into your Python code is easy. Use either of these methods:
Command Palette
Click inside a Python notebook cell or editor and choose Insert Snippet from the Command Palette.
A list of snippets appears. You can type to filter the list; start typing sk to filter the list for snippets provided by this extension. You can further filter the list by continuing to type the desired prefix.
Choose a snippet from the list and it is inserted into your Python code.
Placeholders indicate the minimum arguments required to train a model. Use the tab key to step through the placeholders.
IntelliSense
Start typing sk in a Python notebook cell or editor.
The IntelliSense pop up will appear. You can further filter the pop up list by continuing to type the desired prefix.
Choose a snippet from the pop up and it is inserted into your Python code.
Placeholders indicate the minimum arguments required to train a model. Use the tab key to step through the placeholders.
You can also trigger IntelliSense by typing Ctrl+Space.
2. Features
2.1 Setup
The following snippets are triggered by sk-setup and provide the starting point for creating models. Usually inserted near the beginning of a code file or notebook, sk-setup is the key snippet for importing modules that are commonly used in the machine learning workflow, and setting defaults that apply to data visualizations.
| Snippet |
Placeholders |
Description |
sk-setup |
pio.renderers.default
pio.templates.default |
Provides the initial starting point for creating models. Imports commonly used modules (pandas, numpy, json, pickle, plotly.express, and plotly.io), and sets the default figure renderer and template. |
2.2 Read training data
The following snippets are triggered by sk-read and provide features for creating pandas data frames from Comma Separated Value (.csv), Microsoft Excel (.xlsx), Feather (.feather), and Parquet (.parquet) format files. Tabular data stored in pandas data frames is a common source of training data required for fitting scikit-learn models.
| Snippet |
Placeholders |
Description |
sk-read-csv |
df,
file |
Read tabular training data from CSV (.csv) file (file) into pandas data frame (df) and report info. |
sk-read-excel |
df,
file |
Read tabular training data from Excel (.xlsx) file (file) into pandas data frame (df) and report info. |
sk-read-feather |
df,
file |
Read tabular training data from Feather (.feather) file (file) into pandas data frame (df) and report info. |
sk-read-parquet |
df,
file |
Read tabular training data from Parquet (.parquet) file (file) into pandas data frame (df) and report info. |
2.3 Preprocessing for supervised learning
The following snippets are triggered by sk-prep-target and provide features for preparing tabular training data for supervised learning. The data preparation process involves extracting one or more named features (X) and a named target variable (y) from an input data frame (df), and creating an input training dataset. The data arrays are used to train models for a range of supervised machine learning tasks.
Optionally, the process can also include extracting one or more secondary variables (Z) that are used to better understand the results of model fitting. It is important to note that these secondary variables do not play any role in the model fitting itself, but they can be useful for interpreting the results.
| Snippet |
Placeholders |
Description |
sk-prep-target-features |
X1, X2, X3,
Y
df,
X, y |
Prepare training data (X, y) for supervised learning. Training data is identified by a sequence of feature names (X1, X2, X3, ...), and a target name (Y), sourced from a data frame (df). |
sk-prep-target-features-secondary |
X1, X2, X3,
Y
Z1, Z2, Z3,
df,
X, y, Z |
Prepare training data (X, y) for supervised learning, and prepare secondary data (Z) for model evaluation. Training data is identified by a sequence of feature names (X1, X2, X3, ...), and a target name (Y), sourced from a data frame (df). Secondary data is also identified by a sequence of feature names (Z1, Z2, Z3, ...) sourced from the same data frame (df).
Note: Secondary data is used only for interpreting model output and plays no role in model training. |
sk-prep-train_test_split |
X, y,
⚙ train_size,
⚙ random_state |
Randomly split input data (X, y) into training and test sets using the train_test_split function with the supplied parameters (⚙). Holding out a test set from the training process helps to evaluate the performance of supervised learning models. |
2.4 Preprocessing for unsupervised learning
The following snippets are triggered by sk-prep-features and provide features for preparing tabular training data for unsupervised learning. The data preparation process involves extracting one or more named features (X) from an input data frame (df), and creating an input training dataset. The data array is used to train models for a range of unsupervised machine learning tasks. In contrast to supervised learning, there is no target variable (y) in unsupervised learning.
Similar to the supervised learning case, the process can optionally include extracting one or more secondary variables (Z) that are used to better understand the results of model fitting. It is important to note that these secondary variables do not play any role in the model fitting itself, but they can be useful for interpreting the results.
| Snippet |
Placeholders |
Description |
sk-prep-features |
X1, X2, X3
df,
X |
Prepare training data features (X) for unsupervised learning. Training data is identified by a sequence of feature names (X1, X2, X3, ...) sourced from a data frame (df). |
sk-prep-features-secondary |
X1, X2, X3,
Z1, Z2, Z3,
df,
X, Z |
Prepare training data features (X) for unsupervised learning, and prepare secondary data (Z) for model evaluation. Training data is identified by a sequence of feature names (X1, X2, X3, ...) sourced from a data frame (df). Secondary data is also identified by a sequence of feature names (Z1, Z2, Z3, ...) sourced from the same data frame (df).
Note: Secondary data is used only for interpreting model output and plays no role in model training. |
2.5 Regression
2.5.1 Linear regression
The following snippets are triggered by sk-regress-linear and provide features for various types of linear regression ranging from simple ordinary least squares (LinearRegression), regression with a transformed target (TransformedTargetRegressor), regression with transformed features (FunctionTransformer, PolynomialFeatures, SplineTransformer), and regularized models such as ridge regression (Ridge), lasso regression (Lasso), and elastic net regression (ElasticNet).
| Snippet |
Placeholders |
Description |
sk-regress-linear |
estimator_linear,
⚙ fit_intercept,
⚙ positive,
X, y |
Linear regression: Create and fit a LinearRegression regression model (estimator_linear) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-transform-target |
estimator_transform_target,
⚙ func,
⚙ func_inverse,
⚙ fit_intercept,
⚙ positive,
⚙ check_inverse,
X, y |
Linear regression with transformed target: Create and fit a TransformedTargetRegressor regression model (estimator_transform_target) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-transform |
estimator_transform,
⚙ func,
⚙ fit_intercept,
⚙ positive,
X, y |
Linear regression with transformed features: Create and fit a LinearRegression with FunctionTransformer regression model (estimator_transform) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-polynomial |
estimator_polynomial,
⚙ degree,
⚙ fit_intercept,
⚙ positive,
X, y |
Polynomial regression: Create and fit a LinearRegression with PolynomialFeatures regression model (estimator_polynomial) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-spline |
estimator_spline,
⚙ n_knots,
⚙ degree,
⚙ knots,
⚙ extrapolation,
⚙ fit_intercept,
⚙ positive,
X, y |
Spline regression: Create and fit a LinearRegression with SplineTransformer regression model (estimator_spline) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-pcr |
estimator_pcr,
⚙ n_components,
⚙ whiten,
⚙ fit_intercept,
⚙ positive,
X, y |
Principal component regression (PCR): Create and fit a LinearRegression with PCA regression model (estimator_pcr) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-pls |
estimator_pls,
⚙ n_components,
⚙ scale,
⚙ max_iter,
X, y |
Partial least squares regression (PLS): Create and fit a PLSRegression regression model (estimator_pls) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-ridge |
estimator_ridge,
⚙ alpha,
⚙ fit_intercept,
⚙ positive,
X, y |
Ridge regression: Create and fit a Ridge regression model (estimator_ridge) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-ridgecv |
estimator_ridgecv,
⚙ alphas,
⚙ cv,
⚙ fit_intercept,
X, y |
Ridge regression with cross-validation: Create and fit a RidgeCV regression model (estimator_ridgecv) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-lasso |
estimator_lasso,
⚙ alpha,
⚙ fit_intercept
⚙ positive,
⚙ selection,
⚙ random_state,
X, y |
Lasso regression: Create and fit a Lasso regression model (estimator_lasso) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-lassocv |
estimator_lassocv,
⚙ alphas,
⚙ cv,
⚙ fit_intercept,
⚙ positive,
⚙ selection,
⚙ random_state,
X, y |
Lasso regression with cross-validation: Create and fit a LassoCV regression model (estimator_lassocv) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-elasticnet |
estimator_elasticnet,
⚙ alpha,
⚙ l1_ratio,
⚙ fit_intercept,
⚙ positive,
⚙ selection,
⚙ random_state,
X, y |
ElasticNet regression: Create and fit an ElasticNet regression model (estimator_elasticnet) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-linear-elasticnetcv |
estimator_elasticnetcv,
⚙ l1_ratio,
⚙ alphas,
⚙ fit_intercept,
⚙ positive,
⚙ selection,
⚙ random_state,
X, y |
ElasticNet regression with cross-validation: Create and fit an ElasticNetCV regression model (estimator_elasticnetcv) with the supplied parameters (⚙) and training data (X, y). |
2.5.2 Nearest neighbour regression
The following snippets are triggered by sk-regress-neighbors and provide features for nearest neighbour based regression. These non-parametric methods make predictions from the local neighbourhood of each query sample in the feature space, allowing the model to adapt naturally to complex relationships without assuming a specific functional form.
| Snippet |
Placeholders |
Description |
sk-regress-neighbors-k |
estimator_knn,
⚙ n_neighbors,
⚙ weights,
⚙ algorithm,
⚙ leaf_size,
⚙ p,
⚙ metric,
X, y |
K-nearest neighbors regression: Create and fit a KNeighborsRegressor model (estimator_knn) with preprocessing pipeline using the supplied parameters (⚙) and training data (X, y). Uses make_pipeline with StandardScaler for feature scaling. |
sk-regress-neighbors-radius |
estimator_radius,
⚙ radius,
⚙ weights,
⚙ algorithm,
⚙ leaf_size,
⚙ p,
⚙ metric,
X, y |
Radius neighbors regression: Create and fit a RadiusNeighborsRegressor model (estimator_radius) with preprocessing pipeline using the supplied parameters (⚙) and training data (X, y). Uses make_pipeline with StandardScaler for feature scaling. |
2.5.3 Gaussian process regression
The following snippets are triggered by sk-regress-gaussian and provide features for Gaussian Process Regression (GPR). GPR is a non-parametric Bayesian approach that models uncertainty in predictions and uses kernels to capture complex structure.
| Snippet |
Placeholders |
Description |
|
sk-regress-gaussian-process |
estimator_gp,
⚙ kernel,
⚙ alpha,
⚙ n_restarts_optimizer,
⚙ random_state,
X, y |
Gaussian Process Regression: Create and fit a GaussianProcessRegressor model (estimator_gp) with the supplied parameters (⚙) and training data (X, y). |
|
sk-regress-gaussian-process-kernel |
estimator_gp_kernel,
⚙ kernel,
⚙ alpha,
⚙ n_restarts_optimizer,
⚙ random_state,
X, y |
Gaussian Process Regression with custom kernel: Create and fit a GaussianProcessRegressor model (estimator_gp_kernel) with a custom kernel, the supplied parameters (⚙) and training data (X, y).
Build kernels from components in sklearn.gaussian_process.kernels (e.g., RBF, Matern, RationalQuadratic, DotProduct, ExpSineSquared, WhiteKernel, ConstantKernel) and combine them with + (additive) and * (multiplicative) operators to encode structure. Example: ConstantKernel(1.0, (1e-3, 1e3)) * RBF(1.0, (1e-3, 1e3)) + WhiteKernel(1e-5, (1e-8, 1e1)). Set hyperparameter bounds (e.g., length_scale_bounds) to enable automatic tuning via log-marginal-likelihood optimization.
See Kernels for Gaussian Processes for more details. |
. |
sk-regress-gaussian-transform-target |
estimator_gp_transform_target,
⚙ func,
⚙ func_inverse,
⚙ kernel,
⚙ alpha,
⚙ n_restarts_optimizer,
⚙ random_state,
X, y |
Gaussian Process Regression with transformed target: Create and fit a TransformedTargetRegressor wrapping a GaussianProcessRegressor (estimator_gp_transform_target) with the supplied parameters (⚙) and training data (X, y).
Transforming the target (y) to approximate a Normal distribution honours a key assumption of Gaussian process regression and can improve predictive performance. |
|
2.5.4 Ensemble regression
The following snippets are triggered by sk-regress-ensemble and provide features for various types of ensemble regression. Ensemble regression models combine multiple base regression models to create a model that has superior performance compared to a single base model. Rather than relying on a single model's prediction, ensemble methods aggregate predictions from several models to produce a final result that is typically more accurate.
| Snippet |
Placeholders |
Description |
sk-regress-ensemble-random-forest |
estimator_random_forest,
⚙ n_estimators,
⚙ criterion,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Random forest regression: Create and fit a RandomForestRegressor regression model (estimator_random_forest) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-ensemble-extra-trees |
estimator_extra_trees,
⚙ n_estimators,
⚙ criterion,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Extremely randomized trees (extra-trees) regression: Create and fit an ExtraTreesRegressor regression model (estimator_extra_trees) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-ensemble-gradient-boosting |
estimator_gradient_boosting,
⚙ n_estimators,
⚙ loss,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Gradient boosting regression: Create and fit a GradientBoostingRegressor regression model (estimator_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-ensemble-hist-gradient-boosting |
estimator_hist_gradient_boosting,
⚙ loss,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Histogram-based gradient boosting regression: Create and fit a HistGradientBoostingRegressor regression model (estimator_hist_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-ensemble-stacking |
estimator_stacking,
⚙ estimators,
⚙ final_estimator,
⚙ cv,
⚙ passthrough,
X, y |
Stack of estimators with a final regressor: Create and fit a StackingRegressor regression model (estimator_stacking) with the supplied parameters (⚙) and training data (X, y).
Note: By default, all regression estimators in the current scope are collected for stacking. |
sk-regress-ensemble-voting |
estimator_voting,
⚙ estimators,
⚙ weights,
X, y |
Voting regression: Create and fit a VotingRegressor regression model (estimator_voting) with the supplied parameters (⚙) and training data (X, y).
Note: By default, all regression estimators in the current scope are collected for voting. |
2.5.5 Regression helpers
The following snippets are triggered by sk-regress and provide features for regression helpers.
| Snippet |
Placeholders |
Description |
sk-regress-dummy |
estimator_dummy,
⚙ strategy,
⚙ constant,
⚙ quantile,
X, y |
Dummy regression: Create and fit a DummyRegressor regression model (estimator_dummy) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-report |
estimator,
X,
y_true,
y_pred
|
Regression report: Apply a regression model (estimator) to an input dataset (X) to predict values (y_pred) using the model's predict() function and compare the results with the true values (y_true). Print diagnostic information with the mean_squared_error() and r2_score() functions to evaluate the performance of the regressor on the supplied data. |
2.6 Quantile regression
The following snippets are triggered by sk-regress-quantile and provide features for various types of quantile regression.
In contrast to other regression types (sk-regress-linear, sk-regress-neighbors, sk-regress-gaussian, and sk-regress-ensemble), which estimate the conditional mean of the target variable (y) given the features (X), quantile regression estimates conditional quantiles, these are values that split the distribution of the target variable (y) at specific probability levels given the features (X). Conditional quantiles provide a richer chararcterization of the target distribution than the conditional mean and can reveal if a predictor impacts the low, middle, or high end of outcomes differently. Conditional quantiles are also useful for identifying outliers or modelling a skewed target variable.
Quantile regression snippets are further subdivided into sk-regress-quantile-linear which provides features for various types of linear quantile regression, and sk-regress-quantile-ensemble which provides features for ensemble quantile regression.
2.6.1 Linear quantile regression
The following snippets are triggered by sk-regress-quantile-linear and provide features for various types of linear quantile regression ranging from basic quantile regression (QuantileRegressor), to quantile regression with a transformed target (TransformedTargetRegressor), and quantile regression with transformed features (FunctionTransformer, PolynomialFeatures, SplineTransformer).
| Snippet |
Placeholders |
Description |
sk-regress-quantile-linear |
estimator_quantile_linear,
⚙ quantile,
⚙ alpha,
⚙ fit_intercept,
⚙ solver,
X, y |
Quantile regression: Create and fit a QuantileRegressor regression model (estimator_quantile_linear) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-linear-transform-target |
estimator_quantile_transform_target,
⚙ func,
⚙ func_inverse,
⚙ quantile,
⚙ alpha,
⚙ fit_intercept,
⚙ solver,
⚙ check_inverse,
X, y |
Quantile regression with transformed target: Create and fit a TransformedTargetRegressor wrapping a QuantileRegressor regression model (estimator_quantile_transform_target) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-linear-transform |
estimator_quantile_transform,
⚙ func,
⚙ quantile,
⚙ alpha,
⚙ fit_intercept,
⚙ solver,
X, y |
Quantile regression with transformed features: Create and fit a QuantileRegressor with FunctionTransformer regression model (estimator_quantile_transform) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-linear-polynomial |
estimator_quantile_polynomial,
⚙ degree,
⚙ quantile,
⚙ alpha,
⚙ fit_intercept,
⚙ solver,
X, y |
Quantile polynomial regression: Create and fit a QuantileRegressor with PolynomialFeatures regression model (estimator_quantile_polynomial) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-linear-spline |
estimator_quantile_spline,
⚙ n_knots,
⚙ degree,
⚙ knots,
⚙ extrapolation,
⚙ quantile,
⚙ alpha,
⚙ fit_intercept,
⚙ solver,
X, y |
Quantile spline regression: Create and fit a QuantileRegressor with SplineTransformer regression model (estimator_quantile_spline) with the supplied parameters (⚙) and training data (X, y). |
2.6.2 Ensemble quantile regression
The following snippets are triggered by sk-regress-quantile-ensemble and provide features for various types of ensemble quantile regression including adaptations of the scikit-learn built-in types GradientBoostingRegressor and HistGradientBoostingRegressor for quantile loss, as well as RandomForestQuantileRegressor and ExtraTreesQuantileRegressor from the quantile-forest package.
Predicting multiple quantiles: Among the quantile regression ensemble methods, RandomForestQuantileRegressor and ExtraTreesQuantileRegressor can predict multiple quantiles after fitting, allowing you to specify different quantiles at prediction time via the predict() method's quantiles parameter. In contrast, GradientBoostingRegressor and HistGradientBoostingRegressor require you to specify the target quantile during model creation (via the alpha or quantile parameter), and each fitted model can only predict that single quantile.
| Snippet |
Placeholders |
Description |
sk-regress-quantile-ensemble-random-forest |
estimator_quantile_random_forest,
⚙ n_estimators,
⚙ default_quantiles,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Quantile Random Forest: Create and fit a RandomForestQuantileRegressor regression model (estimator_quantile_random_forest) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-ensemble-extra-trees |
estimator_quantile_extra_trees,
⚙ n_estimators,
⚙ default_quantiles,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Quantile Extremely Randomized Trees: Create and fit an ExtraTreesQuantileRegressor regression model (estimator_quantile_extra_trees) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-ensemble-gradient-boosting |
estimator_quantile_gradient_boosting,
⚙ n_estimators,
⚙ alpha,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Quantile Gradient Boosting: Create and fit a GradientBoostingRegressor regression model (estimator_quantile_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-ensemble-hist-gradient-boosting |
estimator_quantile_hist_gradient_boosting,
⚙ quantile,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Quantile histogram-based gradient boosting: Create and fit a HistGradientBoostingRegressor regression model (estimator_quantile_hist_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
2.6.3 Quantile regression helpers
The following snippets are triggered by sk-regress-quantile and provide features for quantile regression helpers.
| Snippet |
Placeholders |
Description |
sk-regress-quantile-dummy |
estimator_quantile_dummy,
⚙ quantile,
X, y |
Dummy quantile regression: Create and fit a DummyRegressor model for quantile regression (estimator_quantile_dummy) with the supplied parameters (⚙) and training data (X, y). |
sk-regress-quantile-report |
estimator_quantile,
X,
y_true,
y_pred
|
Quantile regression report: Apply a quantile regression model (estimator_quantile) to an input dataset (X) to predict values (y_pred) using the model's predict() function and compare the results with the true values (y_true). Print diagnostic information with the d2_pinball_score() function to evaluate the performance of the quantile regressor on the supplied data. |
2.7 Classification
The following snippets are triggered by sk-classify and provide features for various types of classification models. Classification is a supervised learning task where the goal is to predict discrete class labels for input data. These models learn from labeled training data and can be used for binary classification (two classes) or multi-class classification problems.
2.7.1 Linear classification
The following snippets are triggered by sk-classify-linear and provide features for various types of linear classification models. Linear classifiers separate classes using linear decision boundaries in the feature space. These models are often computationally efficient, interpretable, and serve as good baseline models for many classification problems. Common approaches include linear discriminant analysis, logistic regression, and linear support vector machines.
| Snippet |
Placeholders |
Description |
sk-classify-linear-lda |
estimator_lda,
⚙ solver,
⚙ shrinkage,
⚙ n_components,
⚙ store_covariance,
X, y |
Linear Discriminant Analysis (LDA): Create and fit a LinearDiscriminantAnalysis classification model (estimator_lda) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-linear-qda |
estimator_qda,
⚙ priors,
⚙ reg_param,
⚙ store_covariance,
X, y |
Quadratic Discriminant Analysis (QDA): Create and fit a QuadraticDiscriminantAnalysis classification model (estimator_qda) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-linear-logistic |
estimator_logistic,
⚙ C,
⚙ penalty,
⚙ solver,
⚙ multi_class,
⚙ max_iter,
⚙ random_state,
X, y |
Logistic Regression: Create and fit a LogisticRegression classification model (estimator_logistic) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-linear-svm |
estimator_svm,
⚙ C,
⚙ penalty,
⚙ loss,
⚙ dual,
⚙ max_iter,
⚙ random_state,
X, y |
Linear Support Vector Machine: Create and fit a LinearSVC classification model (estimator_svm) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-linear-svm-sgd |
estimator_sgd,
⚙ loss,
⚙ penalty,
⚙ alpha,
⚙ learning_rate,
⚙ eta0,
⚙ max_iter,
⚙ random_state,
X, y |
SGD Classifier: Create and fit a SGDClassifier classification model (estimator_sgd) with the supplied parameters (⚙) and training data (X, y). Uses stochastic gradient descent for fast training on large datasets. |
sk-classify-linear-perceptron |
estimator_perceptron,
⚙ penalty,
⚙ alpha,
⚙ max_iter,
⚙ random_state,
X, y |
Perceptron: Create and fit a Perceptron classification model (estimator_perceptron) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-linear-ridge |
estimator_ridge,
⚙ alpha,
⚙ solver,
⚙ max_iter,
⚙ random_state,
X, y |
Ridge Classifier: Create and fit a RidgeClassifier classification model (estimator_ridge) with the supplied parameters (⚙) and training data (X, y). |
2.7.2 Nearest neighbour classification
The following snippets are triggered by sk-classify-neighbors and provide features for nearest neighbour based classification. These non-parametric methods make predictions from the local neighbourhood of each query sample in the feature space, allowing the model to adapt naturally to complex relationships without assuming a specific functional form.
| Snippet |
Placeholders |
Description |
sk-classify-neighbors-k |
estimator_knn,
⚙ n_neighbors,
⚙ weights,
⚙ algorithm,
⚙ leaf_size,
⚙ p,
⚙ metric,
X, y |
K-nearest neighbors classification: Create and fit a KNeighborsClassifier model with preprocessing pipeline using the supplied parameters (⚙) and training data (X, y). Uses make_pipeline with StandardScaler for proper feature scaling. |
sk-classify-neighbors-radius |
estimator_radius,
⚙ radius,
⚙ weights,
⚙ algorithm,
⚙ leaf_size,
⚙ p,
⚙ metric,
⚙ outlier_label,
X, y |
Radius neighbors classification: Create and fit a RadiusNeighborsClassifier model with preprocessing pipeline using the supplied parameters (⚙) and training data (X, y). Uses make_pipeline with StandardScaler for proper feature scaling. |
sk-classify-neighbors-centroid |
estimator_centroid,
⚙ metric,
⚙ shrink_threshold,
⚙ priors
X, y |
Nearest centroid classification: Create and fit a NearestCentroid model with preprocessing pipeline using the supplied parameters (⚙) and training data (X, y). Uses make_pipeline with StandardScaler for proper feature scaling. |
2.7.3 Naive Bayes classification
The following snippets are triggered by sk-classify-bayes and provide features for Naive Bayes classification algorithms. These are fast, simple generative classifiers that assume conditional independence of features given the class. Each variant has specific assumptions about the feature distribution and format; choose preprocessing that matches those assumptions rather than generic feature scaling.
| Snippet |
Placeholders |
Description |
sk-classify-bayes-gaussian |
estimator_gaussian_nb,
⚙ priors,
⚙ var_smoothing,
X, y |
Gaussian Naive Bayes: Create and fit a GaussianNB classification model (estimator_gaussian_nb) with the supplied parameters (⚙) and training data (X, y). Intended for continuous features; standardization is not required. For skewed/heavy‑tailed features consider PowerTransformer or QuantileTransformer rather than StandardScaler. |
sk-classify-bayes-multinomial |
estimator_multinomial_nb,
⚙ alpha,
⚙ fit_prior,
⚙ class_prior,
X, y |
Multinomial Naive Bayes: Create and fit a MultinomialNB model (estimator_multinomial_nb) with the supplied parameters (⚙) and training data (X, y). Expects non‑negative features (e.g., counts or tf‑idf). For text, precede with CountVectorizer/TfidfVectorizer instead of StandardScaler. |
sk-classify-bayes-bernoulli |
estimator_bernoulli_nb,
⚙ alpha,
⚙ binarize,
⚙ fit_prior,
⚙ class_prior,
X, y |
Bernoulli Naive Bayes: Create and fit a BernoulliNB model (estimator_bernoulli_nb) with the supplied parameters (⚙) and training data (X, y). Designed for binary (0/1) features. If inputs are continuous, set binarize or add a Binarizer step—do not scale. |
sk-classify-bayes-complement |
estimator_complement_nb,
⚙ alpha,
⚙ fit_prior,
⚙ class_prior,
⚙ norm,
X, y |
Complement Naive Bayes: Create and fit a ComplementNB model (estimator_complement_nb) with the supplied parameters (⚙) and training data (X, y). More robust for imbalanced text; requires non‑negative features (counts/tf‑idf). |
sk-classify-bayes-categorical |
estimator_categorical_nb,
⚙ alpha,
⚙ fit_prior,
⚙ class_prior,
⚙ min_categories,
X, y |
Categorical Naive Bayes: Create and fit a CategoricalNB model (estimator_categorical_nb) with the supplied parameters (⚙) and training data (X, y). Expects integer‑encoded categorical features; use OrdinalEncoder with appropriate handling of unknowns. Do not scale. |
2.7.4 Ensemble classification
The following snippets are triggered by sk-classify-ensemble and provide features for various types of ensemble classification. Ensemble classification models combine multiple base classification models to create a more robust and accurate classifier. These methods typically improve classification performance by aggregating predictions from several models, reducing overfitting, and increasing the model's ability to generalize to new data. Common ensemble techniques include bagging, boosting, and stacking approaches.
| Snippet |
Placeholders |
Description |
sk-classify-ensemble-random-forest |
estimator_random_forest,
⚙ n_estimators,
⚙ criterion,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Random forest classification: Create and fit a RandomForestClassifier classification model (estimator_random_forest) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-ensemble-extra-trees |
estimator_extra_trees,
⚙ n_estimators,
⚙ criterion,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Extremely randomized trees (extra-trees) classification: Create and fit an ExtraTreesClassifier classification model (estimator_extra_trees) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-ensemble-gradient-boosting |
estimator_gradient_boosting,
⚙ n_estimators,
⚙ loss,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Gradient boosting classification: Create and fit a GradientBoostingClassifier classification model (estimator_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-ensemble-hist-gradient-boosting |
estimator_hist_gradient_boosting,
⚙ loss,
⚙ learning_rate,
⚙ min_samples_leaf,
⚙ random_state,
X, y |
Histogram-based gradient boosting classification: Create and fit a HistGradientBoostingClassifier classification model (estimator_hist_gradient_boosting) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-ensemble-stacking |
estimator_stacking,
⚙ estimators,
⚙ final_estimator,
⚙ cv,
⚙ stack_method,
⚙ passthrough,
X, y |
Stack of estimators with a final classifier: Create and fit a StackingClassifier classification model (estimator_stacking) with the supplied parameters (⚙) and training data (X, y).
Note: By default, all classification estimators in the current scope are collected for stacking. |
sk-classify-ensemble-voting |
estimator_voting,
⚙ estimators,
⚙ voting,
⚙ weights,
X, y |
Voting classification: Create and fit a VotingClassifier classification model (estimator_voting) with the supplied parameters (⚙) and training data (X, y).
Note: By default, all classification estimators in the current scope are collected for voting. |
2.7.5 Classification helpers
The following snippets are triggered by sk-classify and provide features for classification helpers.
| Snippet |
Placeholders |
Description |
sk-classify-dummy |
estimator_dummy,
⚙ strategy,
⚙ random_state,
⚙ constant,
X, y |
Dummy classification: Create and fit a DummyClassifier classification model (estimator_dummy) with the supplied parameters (⚙) and training data (X, y). |
sk-classify-report |
estimator,
X,
y_true,
y_pred
|
Classification report: Apply a classification model (estimator) to an input dataset (X) to predict labels (y_pred) using the model's predict() function and compare the results with the true labels (y_true). Print diagnostic information with the classification_report() and confusion_matrix() functions to evaluate the performance of the classifier on the supplied data. |
|
|
|
2.8 Clustering
The following snippets are triggered by sk-cluster and provide features for various types of clustering models. Clustering is an unsupervised learning task that groups similar data points together based on their features. These models identify natural groupings within a dataset without requiring labeled examples, making them useful for discovering patterns, segmenting data, and identifying structure in unlabeled datasets.
| Snippet |
Placeholders |
Description |
sk-cluster-kmeans |
estimator_kmeans,
⚙ n_clusters,
⚙ init,
⚙ n_init,
⚙ max_iter,
⚙ tol,
⚙ random_state,
⚙ algorithm,
X |
K-means clustering: Create and fit a KMeans clustering model (estimator_kmeans) with the supplied parameters (⚙) and training data (X). Includes StandardScaler in pipeline for proper feature scaling. |
sk-cluster-kmeans-minibatch |
estimator_kmeans_minibatch,
⚙ n_clusters,
⚙ init,
⚙ batch_size,
⚙ max_iter,
⚙ max_no_improvement,
⚙ tol,
⚙ random_state,
⚙ reassignment_ratio,
X |
MiniBatch K-means clustering: Create and fit a MiniBatchKMeans clustering model (estimator_kmeans_minibatch) with the supplied parameters (⚙) and training data (X). Optimized for large datasets with StandardScaler in pipeline. |
sk-cluster-meanshift |
estimator_meanshift,
⚙ bandwidth,
⚙ seeds,
⚙ bin_seeding,
⚙ min_bin_freq,
⚙ cluster_all,
⚙ max_iter,
X |
Mean shift clustering: Create and fit a MeanShift clustering model (estimator_meanshift) with the supplied parameters (⚙) and training data (X). Includes StandardScaler in pipeline for proper feature scaling. |
sk-cluster-dbscan |
estimator_dbscan,
⚙ eps,
⚙ min_samples,
⚙ metric,
⚙ algorithm,
⚙ leaf_size,
X |
DBSCAN clustering: Create and fit a DBSCAN clustering model (estimator_dbscan) with the supplied parameters (⚙) and training data (X). Includes StandardScaler in pipeline which is critical for distance-based algorithms. |
sk-cluster-hdbscan |
estimator_hdbscan,
⚙ min_cluster_size,
⚙ min_samples,
⚙ cluster_selection_epsilon,
⚙ cluster_selection_method,
⚙ alpha,
⚙ metric,
⚙ alpha,
⚙ algorithm,
⚙ leaf_size,
X |
HDBSCAN clustering: Create and fit an HDBSCAN clustering model (estimator_hdbscan) with the supplied parameters (⚙) and training data (X). |
sk-cluster-predict |
estimator,
X |
Cluster prediction: Apply a clustering model (estimator) to an input dataset (X) to predict cluster labels using the model's predict() function. Cluster labels are output to a new dataset (X_estimator_cluster). Only available for models that support prediction on new data. |
2.9 Density estimation
The following snippets are triggered by sk-density and provide features for various types of density estimation models. Density estimation is an unsupervised learning task that creates a model of the probability distribution from which the observed data is drawn. These models can be used to generate new samples, detect outliers, and estimate the likelihood of data points.
| Snippet |
Placeholders |
Description |
sk-density-kernel |
estimator_kernel_density,
⚙ bandwidth,
⚙ kernel,
⚙ metric,
X |
Kernel Density Estimation: Create and fit a KernelDensity density estimation model (estimator_kernel_density) with the supplied parameters (⚙) and training data (X). |
sk-density-gaussian-mixture |
estimator_gaussian_mixture,
⚙ n_components,
⚙ covariance_type,
⚙ init_params,
⚙ random_state,
⚙ max_iter,
X |
Gaussian Mixture Model: Create and fit a GaussianMixture density estimation model (estimator_gaussian_mixture) with the supplied parameters (⚙) and training data (X). |
sk-density-sample-kernel |
estimator,
⚙ n_samples,
⚙ random_state, |
Sample from Kernel Density model: Generate random samples from a fitted kernel density model (estimator) using the sample() function with the supplied parameters (⚙). Samples are output to a new dataset (estimator_samples). |
sk-density-sample-gaussian-mixture |
estimator,
⚙ n_samples, |
Sample from Gaussian Mixture model: Generate random samples from a fitted Gaussian Mixture density model (estimator) using the sample() function with the supplied parameters (⚙). Samples are output to new datasets (estimator_samples, and estimator_components). |
sk-density-score-samples |
estimator,
X |
Density score of each sample: Apply a density estimation model (estimator) to an input dataset (X) to evaluate the log-likelihood of each sample using the model's score_samples() function. The log-likelihood is output to a new dataset (X_estimator_density), and is normalized to be a probability density, so the value will be low for high-dimensional data. |
sk-density-score |
estimator,
X |
Density score: Apply a density estimation model (estimator) to an input dataset (X) to evaluate the total log-likelihood of the data in X using the model's score() function. This is normalized to be a probability density, so the value will be low for high-dimensional data. |
2.10 Dimensionality reduction
The following snippets are triggered by sk-embed and provide features for various types of dimensionality reduction or embedding. Dimensionality reduction algorithms transform data from a high-dimensional space into a lower-dimensional representation while preserving the most important structure or information. These techniques are useful for visualization, computational efficiency, and removing redundant features.
Note: The umap package may need to be installed separately using pip install umap-learn.
| Snippet |
Placeholders |
Description |
sk-embed-pca |
estimator_pca,
⚙ n_components,
⚙ whiten
X |
Principal Component Analysis: Create and fit a PCA dimensionality reduction model (estimator_pca) with the supplied parameters (⚙) and training data (X). |
sk-embed-kpca |
estimator_kpca,
⚙ n_components,
⚙ kernel,
⚙ gamma,
⚙ degree,
⚙ coef0
X |
Kernel PCA: Create and fit a KernelPCA dimensionality reduction model (estimator_kpca) with the supplied parameters (⚙) and training data (X). |
sk-embed-lle |
estimator_lle,
⚙ n_components,
⚙ n_neighbors,
⚙ method
X |
Locally Linear Embedding: Create and fit a LocallyLinearEmbedding dimensionality reduction model (estimator_lle) with the supplied parameters (⚙) and training data (X). |
sk-embed-isomap |
estimator_isomap,
⚙ n_components,
⚙ n_neighbors,
⚙ radius,
⚙ p
X |
Isometric Mapping: Create and fit an Isomap dimensionality reduction model (estimator_isomap) with the supplied parameters (⚙) and training data (X). |
sk-embed-mds |
estimator_mds,
⚙ n_components,
⚙ metric,
⚙ n_init,
⚙ random_state,
⚙ normalized_stress
X |
Multidimensional Scaling: Create and fit an MDS dimensionality reduction model (estimator_mds) with the supplied parameters (⚙) and training data (X). |
sk-embed-spectral |
estimator_spectral,
⚙ n_components,
⚙ affinity,
⚙ gamma,
⚙ random_state,
⚙ n_neighbors
X |
Spectral Embedding: Create and fit a SpectralEmbedding dimensionality reduction model (estimator_spectral) with the supplied parameters (⚙) and training data (X). |
sk-embed-tsne |
estimator_tsne,
⚙ n_components,
⚙ perplexity,
⚙ random_state,
⚙ n_iter
X |
t-Distributed Stochastic Neighbour Embedding: Create and fit a TSNE dimensionality reduction model (estimator_tsne) with the supplied parameters (⚙) and training data (X). Embedding is output to X_estimator_tsne. |
sk-embed-nca |
estimator_nca,
⚙ n_components,
⚙ init,
⚙ random_state
X, y |
Neighborhood Components Analysis: Create and fit a NeighborhoodComponentsAnalysis dimensionality reduction model (estimator_nca) with the supplied parameters (⚙) and training data (X, y). |
sk-embed-umap |
estimator_umap,
⚙ n_components,
⚙ metric,
⚙ n_neighbors,
⚙ min_dist,
⚙ random_state,
X |
Uniform Manifold Approximation and Projection: Create and fit a UMAP dimensionality reduction model (estimator_umap) with the supplied parameters (⚙) and training data (X). |
sk-embed-transform |
estimator,
X |
Embedding transform: Apply a dimensionality reduction model (estimator) to an input dataset (X) using the model's transform() function. Embedding is output to a new dataset (X_estimator). |
2.11 Anomaly detection
The following snippets are triggered by sk-anomaly and provide features for various types of anomaly detection models. Anomaly detection is the task of identifying rare items, events, or observations that differ significantly from the majority of the data. These models learn patterns in the data and can detect unusual instances that do not conform to expected behavior.
| Snippet |
Placeholders |
Description |
sk-anomaly-one-class-svm |
estimator_one_class_svm
⚙ kernel,
⚙ gamma,
⚙ nu,
⚙ shrinking,
X |
One-Class Support Vector Machine: Create and fit a OneClassSVM anomaly detection model (estimator_one_class_svm) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-one-class-svm-sgd |
estimator_one_class_svm_sgd
⚙ kernel,
⚙ gamma,
⚙ n_components,
⚙ random_state,
⚙ nu,
⚙ fit_intercept,
⚙ max_iter,
⚙ tol,
⚙ shuffle,
⚙ learning_rate,
⚙ eta0,
X |
One-Class SVM with Stochastic Gradient Descent: Create and fit a SGDOneClassSVM with Nystroem kernel anomaly detection model (estimator_one_class_svm_sgd) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-local-outlier-factor |
estimator_lof
⚙ n_neighbors,
⚙ algorithm,
⚙ leaf_size,
⚙ metric,
⚙ contamination,
⚙ novelty,
X |
Local Outlier Factor: Create and fit a LocalOutlierFactor anomaly detection model (estimator_lof) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-isolation-forest |
estimator_isolation_forest
⚙ n_estimators,
⚙ max_samples,
⚙ contamination,
⚙ max_features,
⚙ bootstrap,
⚙ n_jobs,
⚙ random_state,
X |
Isolation Forest: Create and fit an IsolationForest anomaly detection model (estimator_isolation_forest) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-elliptic-envelope |
estimator_elliptic_envelope
⚙ store_precision,
⚙ assume_centered,
⚙ support_fraction,
⚙ contamination,
⚙ random_state,
X |
Elliptic Envelope (Robust Covariance): Create and fit an EllipticEnvelope anomaly detection model (estimator_elliptic_envelope) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-dbscan |
estimator_dbscan
⚙ eps,
⚙ min_samples,
⚙ metric,
⚙ algorithm,
⚙ leaf_size,
X |
DBSCAN: Create and fit a DBSCAN anomaly detection model (estimator_dbscan) with the supplied parameters (⚙) and training data (X). |
sk-anomaly-predict |
estimator,
X |
Anomaly prediction: Apply an anomaly detection model (estimator) to an input dataset (X) to perform outlier classification using the model's predict() function. Outlier class is output to a new dataset (X_estimator_class) where -1 indicates outliers, and +1 indicates inliers. |
sk-anomaly-score |
estimator,
X |
Anomaly score: Apply an anomaly detection model (estimator) to an input dataset (X) to evaluate the outlier score of each sample using the model's descision_function() function. Outlier score is output to a new dataset (X_estimator_score) where negative scores indicate outliers, and positive scores indicate inliers. |
2.12 Model inspection
The following snippets are triggered by sk-inspect and provide features for inspecting and understanding fitted models. Model inspection tools help data scientists gain insights into how their models make predictions, which features are most important, and how changes in input features affect model outputs. These techniques are essential for explaining model behavior, debugging models, and ensuring that models are behaving as expected.
| Snippet |
Placeholders |
Description |
sk-inspect-partial_dependence |
estimator,
X,
⚙ features,
⚙ percentiles,
⚙ grid_resolution,
⚙ kind |
Partial dependence: Compute the partial dependence of a model (estimator) against an input feature dataset (X) using the partial_dependence function with the supplied parameters (⚙). Output partial dependence curves are returned as a dictionary (estimator_partial). |
sk-inspect-permutation_importance |
estimator,
X, y,
⚙ scoring,
⚙ n_repeats,
⚙ random_state,
⚙ sample_weight,
⚙ max_samples |
Permutation importance: Compute the permutation importance of a model (estimator) against an input dataset (X,y) using the permutation_importance function with the supplied parameters (⚙). Output feature importance metrics are returned as a dictionary (estimator_permutation). |
2.13 Model persistence
2.13.1 Pickle
The following snippets are triggered by sk-io-pickle and provide features for reading and writing scikit-learn models in pickle format. Python's built-in pickle module enables saving and loading trained models in binary format. Loading pickle files from untrusted or unknown sources poses significant security risks, as malicious pickle files could execute arbitrary code. As a result it is strongly recommended to only load pickle files from trusted sources.
| Snippet |
Placeholders |
Description |
sk-io-pickle-read |
file,
estimator |
Read an existing model (estimator) from a (.pickle) format file (file). Report the model type and any fitted parameters. |
sk-io-pickle-write |
file,
estimator |
Write a model (estimator) to a (.pickle) format file (file). |
2.13.2 Skops
The following snippets are triggered by sk-io-skops and provide features for reading and writing scikit-learn models in skops format. The skops package provides a more secure alternative to pickle for saving and loading models. It includes additional security features like model verification and safer deserialization to mitigate potential security risks associated with loading models from untrusted sources.
Note: The skops package may need to be installed separately using pip install skops.
| Snippet |
Placeholders |
Description |
sk-io-skops-read |
file,
estimator |
Read an existing model (estimator) from a (.skops) format file (file). Report the model type and any fitted parameters. |
sk-io-skops-write |
file,
estimator |
Write a model (estimator) to a (.skops) format file (file). |
2.14 Argument Snippets
Many scikit-learn function arguments take their values from extensive option lists. The following snippets are triggered with sk-args and provide features allowing you to easily select valid argument values from lists of available options.
| Snippet |
Placeholders |
Description |
sk-args-random_state |
random_state |
Set the random_state argument for reproducibility in randomized algorithms. This argument is the integer seed number used to initialize random number generation; a randomly chosen value is provided by default. |
sk-args-alphas |
logspace or linspace,
start, stop, num |
Set the alphas argument (a logarithmic or linear sequence of regularization parameters) for cross-validation in RidgeCV, LassoCV, and ElasticNetCV regression. |
sk-args-func |
func |
Set the func argument to a FunctionTransformer from a list of common transformations. For use in regression models with transformed features (X), see sk-regress-linear-transform. |
sk-args-func-inverse |
func, inverse_func |
Set the func and inverse_func arguments to a FunctionTransformer from a list of common forward and inverse transformation function pairs. For use in regression models with transformed target (y), see sk-regress-linear-transform-target. |
sk-args-spline-extrapolation |
extrapolation |
Set the SplineTransformer extrapolation behavior beyond the minimum and maximum of the training data, see sk-regress-linear-spline. |
sk-args-kernel |
kernel |
Set the kernel type for kernel density estimation from an option list. You can provide this argument when creating a KernelDensity model, see sk-density-kernel. |
|
|
|
3. Release Notes
3.1 Python packages
3.1.1 Scikit-learn
The snippets provided by this extension were developed using scikit-learn version 1.7 but will also produce working Python code for earlier and later versions. Install it before use, for example:
pip install scikit-learn
See the scikit-learn documentation for details.
3.1.2 Quantile-forest
Quantile regression forest snippets (sk-regress-quantile-ensemble-random-forest and sk-regress-quantile-ensemble-extra-trees) depend on the external quantile-forest package, which is not part of scikit-learn. Install it separately before use, for example:
pip install quantile-forest
See the quantile-forest documentation for details.
3.1.3 UMAP
UMAP embedding (sk-embed-umap) depends on the external umap-learn package, which is not part of scikit-learn. Install it separately before use, for example:
pip install umap-learn
See the umap-learn documentation for details.
3.1.4 Skops
The sk-io-skops snippets depend on the external skops package, which is not part of scikit-learn. Install it separately before use, for example:
pip install skops
See the skops documentation for details.
3.2 Using snippets
Editor Support for Snippets
Snippets for producing Python code, including those provided by this extension, are supported in the Python file (.py) editor and in the notebook (.ipynb) editor.
Snippets and IntelliSense
When triggered, the default behaviour of IntelliSense is to show snippets along with other context dependent suggestions. This may result in a long list of suggestions in the IntelliSense pop-up, particularly if the snippet trigger provided by this extension (sk) also matches other symbols in your editor.
It's easy to modify this behaviour using your Visual Studio Code settings. To access the relevant settings go to Preferences > Settings and type snippets in the Search settings field as shown below:
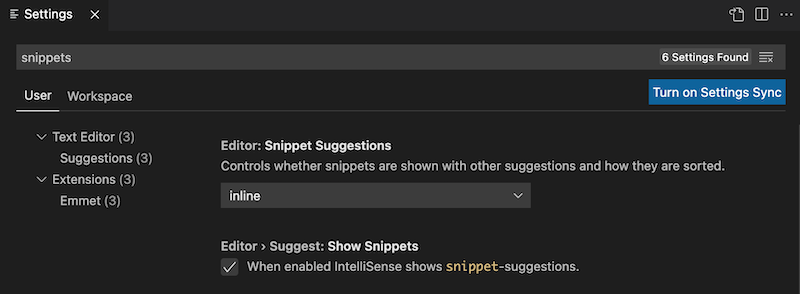
You can control whether snippets are shown with other suggestions and how they are sorted using the Editor: Snippet Suggestions dropdown. Choose one of the options to control how snippet suggestions are shown in the IntelliSense popup:
| Option |
IntelliSense |
top |
Show snippet suggestions on top of other suggestions. |
bottom |
Show snippet suggestions below other suggestions. |
inline |
Show snippet suggestions with other suggestions (default). |
none |
Do not show snippet suggestions. |
You can also use the Editor > Suggest: Show Snippets checkbox to enable or disable snippets in IntelliSense suggestions. When snippets are disabled in IntelliSense they are still accessible through the Command Palette Insert Snippet command.
4. Snippet reference
Snippet prefix triggers are organized in a hierarchical tree structure rooted at sk as shown in the figure below. The snippet hierarchy is designed to ease the user's cognitive load when developing models with this large and complex machine learning package. The branches at the top of the tree outline the main steps in a machine learning workflow, branches at lower levels outline a taxonomy of algorithms for specific tasks, whereas leaf nodes represent particular algorithms. The process of inserting a snippet amounts to navigating the tree and selecting the desired leaf node by either of the methods described in Section 1.3.
sk
├── sk-setup
├── sk-read
│ ├── sk-read-csv
│ ├── sk-read-excel
│ ├── sk-read-feather
│ └── sk-read-parquet
├── sk-prep
│ ├── sk-prep-target-features
│ ├── sk-prep-target-features-secondary
│ ├── sk-prep-train_test_split
│ ├── sk-prep-features
│ └── sk-prep-features-secondary
├── sk-regress
│ ├── sk-regress-linear
│ │ ├── sk-regress-linear
│ │ ├── sk-regress-linear-transform-target
│ │ ├── sk-regress-linear-transform
│ │ ├── sk-regress-linear-polynomial
│ │ ├── sk-regress-linear-spline
│ │ ├── sk-regress-linear-pcr
│ │ ├── sk-regress-linear-pls
│ │ ├── sk-regress-linear-ridge
│ │ ├── sk-regress-linear-ridgecv
│ │ ├── sk-regress-linear-lasso
│ │ ├── sk-regress-linear-lassocv
│ │ ├── sk-regress-linear-elasticnet
│ │ └── sk-regress-linear-elasticnetcv
│ ├── sk-regress-neighbors
│ │ ├── sk-regress-neighbors-k
│ │ └── sk-regress-neighbors-radius
│ ├── sk-regress-gaussian
│ │ ├── sk-regress-gaussian-process
│ │ ├── sk-regress-gaussian-process-kernel
│ │ └── sk-regress-gaussian-transform-target
│ ├── sk-regress-ensemble
│ │ ├── sk-regress-ensemble-random-forest
│ │ ├── sk-regress-ensemble-extra-trees
│ │ ├── sk-regress-ensemble-gradient-boosting
│ │ ├── sk-regress-ensemble-hist-gradient-boosting
│ │ ├── sk-regress-ensemble-stacking
│ │ └── sk-regress-ensemble-voting
│ ├── sk-regress-dummy
│ ├── sk-regress-report
│ └── sk-regress-quantile
│ ├── sk-regress-quantile-linear
│ │ ├── sk-regress-quantile-linear
│ │ ├── sk-regress-quantile-linear-transform-target
│ │ ├── sk-regress-quantile-linear-transform
│ │ ├── sk-regress-quantile-linear-polynomial
│ │ └── sk-regress-quantile-linear-spline
│ ├── sk-regress-quantile-ensemble
│ │ ├── sk-regress-quantile-ensemble-random-forest
│ │ ├── sk-regress-quantile-ensemble-extra-trees
│ │ ├── sk-regress-quantile-ensemble-gradient-boosting
│ │ └── sk-regress-quantile-ensemble-hist-gradient-boosting
│ ├── sk-regress-quantile-dummy
│ └── sk-regress-quantile-report
├── sk-classify
│ ├── sk-classify-linear
│ │ ├── sk-classify-linear-lda
│ │ ├── sk-classify-linear-qda
│ │ ├── sk-classify-linear-logistic
│ │ ├── sk-classify-linear-svm
│ │ ├── sk-classify-linear-svm-sgd
│ │ ├── sk-classify-linear-perceptron
│ │ └── sk-classify-linear-ridge
│ ├── sk-classify-neighbors
│ │ ├── sk-classify-neighbors-k
│ │ ├── sk-classify-neighbors-radius
│ │ └── sk-classify-neighbors-centroid
│ ├── sk-classify-bayes
│ │ ├── sk-classify-bayes-gaussian
│ │ ├── sk-classify-bayes-multinomial
│ │ ├── sk-classify-bayes-bernoulli
│ │ ├── sk-classify-bayes-complement
│ │ └── sk-classify-bayes-categorical
│ ├── sk-classify-ensemble
│ │ ├── sk-classify-ensemble-random-forest
│ │ ├── sk-classify-ensemble-extra-trees
│ │ ├── sk-classify-ensemble-gradient-boosting
│ │ ├── sk-classify-ensemble-hist-gradient-boosting
│ │ ├── sk-classify-ensemble-stacking
│ │ └── sk-classify-ensemble-voting
│ ├── sk-classify-dummy
│ └── sk-classify-report
├── sk-cluster
│ ├── sk-cluster-kmeans
│ ├── sk-cluster-kmeans-minibatch
│ ├── sk-cluster-meanshift
│ ├── sk-cluster-dbscan
│ ├── sk-cluster-hdbscan
│ └── sk-cluster-predict
├── sk-density
│ ├── sk-density-kernel
│ ├── sk-density-gaussian-mixture
│ ├── sk-density-sample-kernel
│ ├── sk-density-sample-gaussian-mixture
│ ├── sk-density-score-samples
│ └── sk-density-score
├── sk-embed
│ ├── sk-embed-pca
│ ├── sk-embed-kpca
│ ├── sk-embed-lle
│ ├── sk-embed-isomap
│ ├── sk-embed-mds
│ ├── sk-embed-spectral
│ ├── sk-embed-tsne
│ ├── sk-embed-nca
│ ├── sk-embed-umap
│ └── sk-embed-transform
├── sk-anomaly
│ ├── sk-anomaly-one-class-svm
│ ├── sk-anomaly-one-class-svm-sgd
│ ├── sk-anomaly-local-outlier-factor
│ ├── sk-anomaly-isolation-forest
│ ├── sk-anomaly-elliptic-envelope
│ ├── sk-anomaly-dbscan
│ ├── sk-anomaly-predict
│ └── sk-anomaly-score
├── sk-inspect
│ ├── sk-inspect-partial_dependence
│ └── sk-inspect-permutation_importance
├── sk-io
│ ├── sk-io-pickle
│ │ ├── sk-io-pickle-read
│ │ └── sk-io-pickle-write
│ └── sk-io-skops
│ ├── sk-io-skops-read
│ └── sk-io-skops-write
└── sk-args
├── sk-args-random_state
├── sk-args-alphas
├── sk-args-func
├── sk-args-func-inverse
├── sk-args-spline-extrapolation
└── sk-args-kernel
Copyright © 2024-2026 Analytic Signal Limited, all rights reserved
| |


